Related Articles
Articles and information about gut microbiomeDean’s Doctor Weekly diary (129): What Vaccines Can’t Prevent
:疫苗不能預防的問題.png)
Dean’s Doctor Weekly diary (129): What Vaccines Can’t Prevent
While the world is eagerly awaiting the early release of the vaccine, have we thought about the many long-term problems that the vaccine cannot prevent?
We have been living a very cautious life for the past half year, cleaning our hands, disinfecting furniture, and eating carefully. There is no denying that personal hygiene can greatly reduce the risk of contracting the new coronavirus and influenza, but an absolutely clean environment can also bring new problems to our health, especially to our children’s development.
According to medical research, environmental hygiene and dietary habits directly affect infants’ gut microbiome, and many childhood health problems such as eczema, asthma, obesity, autism, type 1 diabetes, Crohn’s disease, etc., are closely related to intestinal micro-ecology. It is interesting to note that these problems often occur in “very hygienic” families. As the previous generation had poor environmental hygiene, we often used the excuse of “germs eating germs” to cover up our unhygienic eating habits. Recent studies have pointed out that although the previous generation of children had poor hygiene, they formed a more comprehensive gut flora (microbiome with good diversity), making the immune system more mature and thus reducing the risk of these diseases. The study also found that children who take antibiotics regularly since childhood will increase the risk of obesity, eczema and other immune system diseases, because antibiotics will reduce the diversity of intestinal bacteria, again proving the importance of intestinal flora to health.
Children’s dietary habits are also closely related to the gut microbiome, nowadays many foods with a large number of emulsifier (emulsifier), such as ice cream, fresh milk, salad dressing, etc., in order to make the color and flavor of food more attractive, but research points out that these emulsifiers make the intestinal micro-ecological imbalance (dysbiosis), resulting in intestinal inflammation and decreased immunity, increasing the risk of many chronic diseases.
Improvements in quality of life and environment have apparently reduced certain diseases, such as malnutrition and plague, but they have been replaced by more and more “diseases of affluence”. Under the threat of the new global coronavirus, we have become more concerned about personal hygiene, but living in a very hygienic environment for a long time, we have lost the opportunity of “germs eating bacteria”, and the diverse gut microbiota that maintains our health is thus weakened, and human beings may have to bear the long-term consequences.
In the future, we need to protect and improve the gut microbiome to prevent and cure diseases.
(Originally published in Ming Pao)
Source:https://www.facebook.com/489667057835766/posts/2114955921973530/
Intestinal health and Immunity

Intestine is the important barrier of immunity system. More than 70% of immune cells are concentrated in the intestine to prevent invasion of pathogens. If the intestinal health is reduced, the immunity of oneself will be reduced too.
Therefore, in order to maintain intestinal health, we can start from three fundamental elements, which are water, dietary fiber and probiotics. Coronavirus outbreak is happening in these days and so the citizens are needed to improve their immune ability. Thus, other than drinking more water and doing more exercises, the increase of intake of good bacteria is one of the paramount factors to prevent virus infection! Probiotics are the microbes that improve the hosts’ intestinal health. Intaking of probiotics can maintain the balance of gut microbiome and reduce the toxins produced by the bad bacteria in the gastrointestinal tract. Many molecules involving in the immune system is produced by the good bacteria in the intestine, and it is effectively regulating our immune system.
Lastly, we hope that everyone can stay healthy by maintaining the health of intestine!
Sources: http://www.3phk.com/v5article.asp?id=3473
Gut Microbiota: A Neglected Organ? (5)
Future direction
Enhancements in the big data available on the structure and function of the gut microbiome in healthy and diseased populations will improve our understanding of the role of the gut microbiome in disease genesis and its potential role in therapeutic intervention. For conditions shown to be associated with changes in the microbiota, such as IBD, allergies, obesity and depression, more research is needed to elucidate whether such changes are causative. How can we move from detecting altered microbial community structure to elucidating altered community function? Is there a way to influence the balance of our microbiota, either during its initial establishment in infancy or once it has been established? To date, research has largely focused on bacterial inhabitants of the microbiome. What roles do viruses, fungi and other eukaryotic microbes (such as helminths) play in human health and disease? Can the gut microbiota be manipulated and leveraged for precision medicine? Hopefully, with the collaborative efforts of experts from different specialties, the puzzle of this “neglected organ” will be solved in the near future.
Author: Dr Steven Loo, Professor Tsui Kowk Wing
Gut Microbiota: A Neglected Organ? (4)
What could microbiome analysis data reveal?
Despite the technological advances, our knowledge of the exact structure and function of the human gut microbiome remains limited. The initial activities of the HMP and other microbiome consortia focused on defining the “normal” parameters of the microbial communities in the human body. It was hoped that defining what is normal or ‘healthy’ would facilitate identification of deviations (dysbiosis) from this normal state in various diseases. A healthy gut microbiota composition is highly variable depending on age, ethnicity, lifestyle and dietary habits. However, a certain degree of gut microbiota variation, namely dysbiosis, has implications for intestinal and extra-intestinal disorders. Dysbiosis is often defined as an alteration of gut microbiota composition and a cause or a consequence of the particular disorders. Numerous studies have attempted to establish the dysbiotic state by comparing the structure and function of the microbiome between healthy and disease states. The potential dysbiotic features of the gut microbiome include the beta diversity (bacterial diversity between two groups of study population), the alpha diversity (bacteria diversity in one single individual), and the status of core essential gut microbiota or the proportion of individual signature bacteria, such as Proteobacteria phylum, Faecalibacterium prausnitzii species or Akkermansia muciniphila species.
In inflammatory bowel disease (IBD), for example, differences were seen between the microbiota of affected patients and control subjects without IBD in terms of diversity and proportions of certain signature bacteria, but it was unclear whether these differences were causative or secondary to the presence of the disease. As a result, studies have attempted to characterize the microbiota before the development of the overt disease. Those studies recruited patients at high risk for a specific disease (eg, IBD or type I diabetes) and monitored their microbiota longitudinally, comparing those who subsequently developed disease with those who did not. Other studies examined the effects of faecal transplantation therapy for specific illnesses, such as obesity and diabetes, in an attempt to establish the role of the microbiota in the pathogenesis of the disease. For diseases with well-established animal models, tests of potential causation have been attempted. A study in the UK collected stool samples from four sets of monozygotic twins with weight discrepancies, with the obese twin having a body mass index (BMI) >30 kg/m2 and a sustained BMI difference of ≥5.5 kg/m2 with their co-twin. In this twin cohort study, researchers assumed that the weight discrepancy was dependent on environmental factors alone. Faecal material from “obese” twins and “lean” twins were transplanted into two groups of mice through gastric lavage. Despite consuming the same low-fat low-calorie diet under the controlled experimental environment, mice that received faecal transplants from an “obese” twin became obese, while mice that received faecal transplants from a “lean” twin became lean. This study further illustrated the concept of a causative role for microbiota in disease manifestation.
Author: Dr Steven Loo, Professor Tsui Kwok Wing, Stephen
Gut Microbiota: A Neglected Organ? (3)
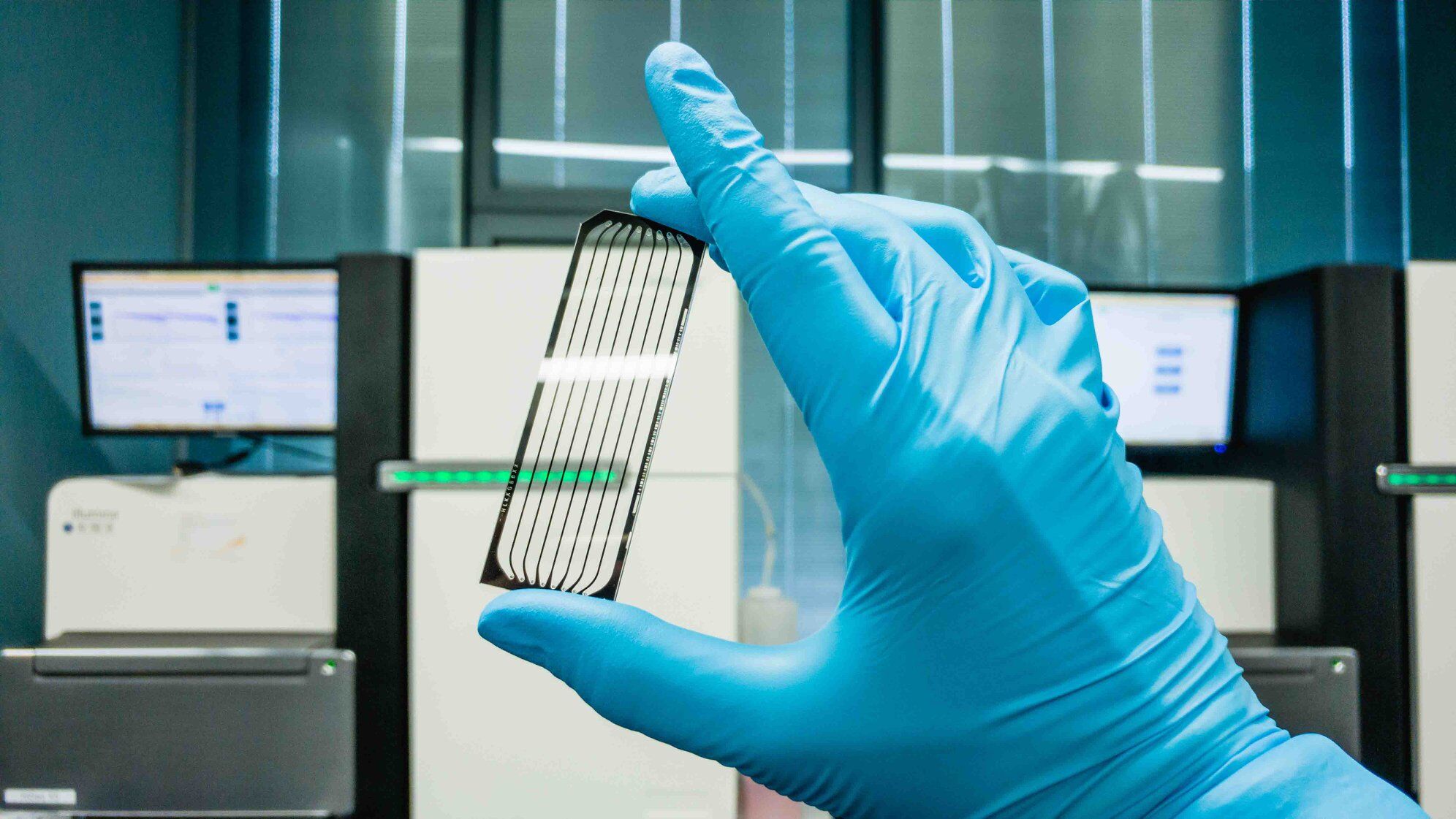
How to analyse the gut microbiome?
For over 100 years, culture-based techniques were the gold standard for classifying microbes. However, these approaches are restricted to the small subset of microbes that survive isolation and specific laboratory growth conditions. Contemporary genomic classification approaches offered a solution to the limitations of culture-based techniques. Unique to prokaryotic organisms, the 16S ribosomal RNA (rRNA) gene is a highly conserved region within the bacterial genome. Interestingly, within the highly conserved 16S rRNA gene are nine hypervariable regions with species-specific signatures. Sequences of the 16S rRNA gene can therefore be compared with the “reference library” for taxonomic classification. The 16S rRNA gene is an ideal and simple tool for profiling gut bacteria.
Standard human gut microbiome studies involve the extraction and sequencing of microbial DNA from a faecal sample collected in a stool container with DNA stabilizer, followed by DNA extraction, gene sequencing and computational bioinformatics analysis (Figure 2). Early human microbial studies relied upon fingerprinting techniques or Sanger sequencing of the amplified 16S rRNA gene. In the past decade, next-generation sequencing (NGS) platforms have offered faster and more efficient sequencing at much lower cost.
16S rRNA analysis is useful for understanding the composition and structural role (anatomy) of microbes in healthy and disease states. However, full assessment of microbial function (physiology) requires other genomic technologies like metagenomic shotgun sequencing. Metagenomics is the analysis of genetic content from the whole microbial ecosystem, and the entire “meta” genome of all microorganisms combined is studied. The term “shotgun sequencing” refers to the generation of sequencing libraries by randomly fragmenting DNA templates of all microbial agents, irrespective of differences between microbial genomes. Metagenomic shotgun sequencing could potentially reveal all the functional genetic content of the microbiota, but the cost and bioinformatics requirement are likely to be much higher than for the usual 16S rRNA analysis. Moreover, if this strategy is employed, there will be significant human DNA contamination in the resulting data. Other ‘omics‘ technologies that could provide insight into the gene expression and metabolic activities of the gut microbiota include metatranscriptomics (RNA expression), metaproteomics (protein) and metabolomics (metabolites). Interestingly, microbial culture remains relevant as an essential tool for functional studies of specific species following ‘omics’ analysis.
